Eigenvalue analysis of Eiffel tower
Eigenvalue analysis of Eiffel tower is done using JuliaFEM 0.3.2 and Julia 0.6. Results show that some code optimization is needed in assembly of global matrices and in particular in storing results to Xdmf.


Mesh details and first eigenmode


Code
using JuliaFEM
using JuliaFEM.Preprocess
using JuliaFEM.Postprocess
# script takes model name as a first argument, e.g.
# model = "EIFFEL_TOWER_TET10_220271", and it is expected
# that there is corresponding mesh file in directory
# eiffel-tower, e.g. "eiffel-tower/EIFFEL_TOWER_TET10_220271.inp"
model = ARGS[1]
mesh = joinpath("eiffel-tower", "$model.inp")
results = "$model"
@timeit to "run performance test" begin
@timeit to "parse input data" begin
mesh = abaqus_read_mesh(mesh)
for (nid, ncoords) in mesh.nodes
mesh.nodes[nid] = 304.8 * ncoords
end
info("element sets = ", collect(keys(mesh.element_sets)))
info("surface sets = ", collect(keys(mesh.surface_sets)))
end
@timeit to "initialize model" begin
tower = Problem(Elasticity, "tower", 3)
tower.elements = create_elements(mesh, "TOWER")
update!(tower, "youngs modulus", 210.0E3)
update!(tower, "poissons ratio", 0.3)
update!(tower, "density", 7.85E-9)
support = Problem(Dirichlet, "fixed", 3, "displacement")
support.elements = create_surface_elements(mesh, "SUPPORT")
update!(support, "geometry", mesh.nodes)
update!(support, "displacement 1", 0.0)
update!(support, "displacement 2", 0.0)
update!(support, "displacement 3", 0.0)
end
@timeit to "solve eigenvalue problem" begin
solver = Solver(Modal, tower, support)
solver.xdmf = Xdmf(results; overwrite=true)
solver.properties.nev = 5
solver.properties.which = :SM
solver()
println("Eigenvalues: ", sqrt(solver.properties.eigvals) / (2*pi))
end
end
print_statistics()
Results
220271 nodes
──────────────────────────────────────────────────────────────────────────────
Time Allocations
────────────────────── ───────────────────────
Tot / % measured: 463s / 86.8% 87.8GiB / 98.8%
Section ncalls time %tot avg alloc %tot avg
──────────────────────────────────────────────────────────────────────────────
run performance test 1 402s 100% 402s 86.7GiB 100% 86.7GiB
solve eigenvalue... 1 395s 98.3% 395s 85.9GiB 99.0% 85.9GiB
save results t... 1 188s 46.8% 188s 13.2GiB 15.2% 13.2GiB
assemble matrices 1 130s 32.4% 130s 49.4GiB 57.0% 49.4GiB
solve eigenval... 1 38.2s 9.50% 38.2s 8.87GiB 10.2% 8.87GiB
parse input data 1 5.39s 1.34% 5.39s 689MiB 0.78% 689MiB
initialize model 1 1.25s 0.31% 1.25s 184MiB 0.21% 184MiB
──────────────────────────────────────────────────────────────────────────────
376120 nodes
──────────────────────────────────────────────────────────────────────────────
Time Allocations
────────────────────── ───────────────────────
Tot / % measured: 745s / 97.3% 155GiB / 99.3%
Section ncalls time %tot avg alloc %tot avg
──────────────────────────────────────────────────────────────────────────────
run performance test 1 725s 100% 725s 154GiB 100% 154GiB
solve eigenvalue... 1 716s 98.7% 716s 152GiB 99.0% 152GiB
save results t... 1 335s 46.2% 335s 23.3GiB 15.1% 23.3GiB
assemble matrices 1 232s 32.0% 232s 87.7GiB 57.1% 87.7GiB
solve eigenval... 1 69.7s 9.61% 69.7s 16.2GiB 10.5% 16.2GiB
parse input data 1 7.72s 1.06% 7.72s 1.20GiB 0.78% 1.20GiB
initialize model 1 1.98s 0.27% 1.98s 324MiB 0.21% 324MiB
──────────────────────────────────────────────────────────────────────────────
921317 nodes
──────────────────────────────────────────────────────────────────────────────
Time Allocations
────────────────────── ───────────────────────
Tot / % measured: 2300s / 99.3% 399GiB / 100%
Section ncalls time %tot avg alloc %tot avg
──────────────────────────────────────────────────────────────────────────────
run performance test 1 2283s 100% 2283s 398GiB 100% 398GiB
solve eigenvalue... 1 2260s 99.0% 2260s 394GiB 99.1% 394GiB
save results t... 1 960s 42.0% 960s 59.9GiB 15.1% 59.9GiB
assemble matrices 1 702s 30.8% 702s 226GiB 56.9% 226GiB
solve eigenval... 1 216s 9.45% 216s 44.6GiB 11.2% 44.6GiB
parse input data 1 18.8s 0.82% 18.8s 2.94GiB 0.74% 2.94GiB
initialize model 1 3.90s 0.17% 3.90s 823MiB 0.20% 823MiB
──────────────────────────────────────────────────────────────────────────────
1327989 nodes
──────────────────────────────────────────────────────────────────────────────
Time Allocations
────────────────────── ───────────────────────
Tot / % measured: 3742s / 100% 600GiB / 100%
Section ncalls time %tot avg alloc %tot avg
──────────────────────────────────────────────────────────────────────────────
run performance test 1 3725s 100% 3725s 599GiB 100% 599GiB
solve eigenvalue... 1 3692s 99.1% 3692s 593GiB 99.1% 593GiB
save results t... 1 1495s 40.1% 1495s 89.3GiB 14.9% 89.3GiB
assemble matrices 1 1181s 31.7% 1181s 339GiB 56.6% 339GiB
solve eigenval... 1 359s 9.63% 359s 71.5GiB 11.9% 71.5GiB
parse input data 1 27.6s 0.74% 27.6s 4.19GiB 0.70% 4.19GiB
initialize model 1 5.14s 0.14% 5.14s 1.19GiB 0.20% 1.19GiB
──────────────────────────────────────────────────────────────────────────────
2357071 nodes
──────────────────────────────────────────────────────────────────────────────
Time Allocations
────────────────────── ───────────────────────
Tot / % measured: 10669s / 100% 1145GiB / 100%
Section ncalls time %tot avg alloc %tot avg
──────────────────────────────────────────────────────────────────────────────
run performance test 1 10653s 100% 10653s 1144GiB 100% 1144GiB
solve eigenvalue... 1 10608s 100% 10608s 1134GiB 99.1% 1134GiB
save results t... 1 3104s 29.1% 3104s 167GiB 14.6% 167GiB
assemble matrices 1 3100s 29.1% 3100s 639GiB 55.9% 639GiB
solve eigenval... 1 1624s 15.2% 1624s 155GiB 13.5% 155GiB
parse input data 1 36.8s 0.35% 36.8s 7.75GiB 0.68% 7.75GiB
initialize model 1 7.64s 0.07% 7.64s 2.22GiB 0.19% 2.22GiB
Further analysis and discussion
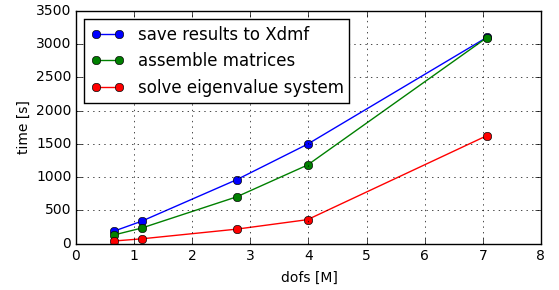
Looks that saving results to Xdmf is taking a long time (should not) and that’s indeed develop target in next releases. Large memory allocation in global assembly is telling that a lot of temporary matrices, so that’s a good development target also in future. From figure that time for storing results and global assembly grows linearly as the function of dofs, while the actual solving of system not. It can be expected that when model is big enough, most of the time is used in solution of system, not in other operations.
